Liu Group
Our group aims to understand the processes cells use to counteract replication stress caused by either internal factors or external factors, and why some regions in the genome are particularly susceptible to those factors.

Liu group aims to understand 1) the measures cells use to counteract replication stress caused by either internal factors (i.e. oncogene activation), or external factors (i.e. folate deficiency), and, 2) why some regions in the genome are particularly susceptible to those factors (e.g. common or rare fragile sites). Specifically, we focus on the two areas shown below. We employ techniques in the fields of cellular and molecular biology, cytogenetics, biochemistry, advanced microscope imaging, whole genome sequencing and mass spectrometry based proteomics.
1) The analysis of mechanisms underlying MiDAS
It is well established that incomplete replication can cause a delay in chromatin condensation that leads to the ‘expression’ of common fragile sites (CFSs) [1, 2]. CFSs are hot spots for deletions and chromosome rearrangements in cancer [3]. We previously have taken part in the discovery of a process called mitotic DNA synthesis (MiDAS) that operates in mitosis is a strategy used by human cells to rescue the incomplete replication at those loci, particularly in cancers cells that has elevated replication stress (RS) [4-7] (Figure 1). Particularly, both RAD52 and POLD3 play a crucial role in MiDAS. Our recent findings demonstrate that: i) RTEL1, a DNA helicase, can prevent the accumulation of G-quadruplex-associated R-loops at difficult-to-replicate loci including CFSs in the human genome in S phase, and can facilitate MiDAS in M phase [8]; ii) both translesion polymerases and replication replication polymerase delta play a crucial role in completing MiDAS [9]. In addition, using a BioID strategy [10], we have identified a panel of factors that could potentially work closely with POLD3 when cells are challenged with RS. We are now investigating the functions of these factors and their relevance to MiDAS.
2) The analysis of folate deficiency induced genome instability
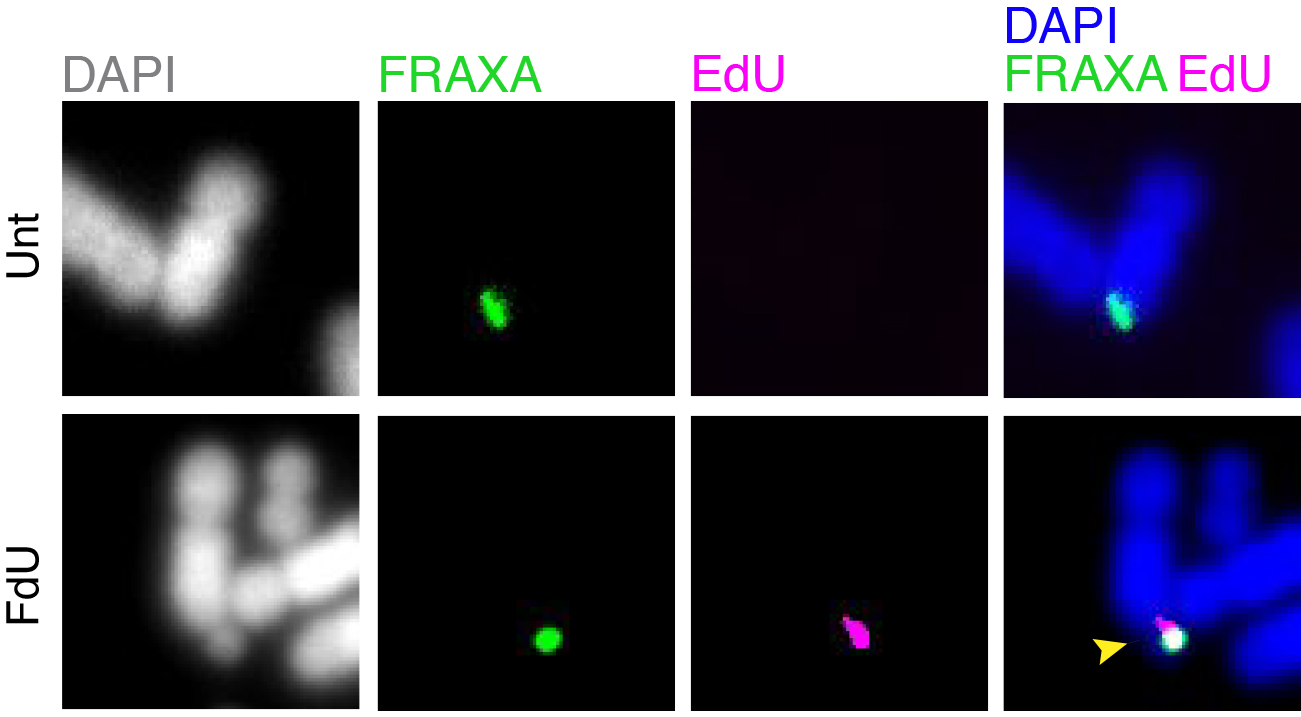
Folate deficiency is known to be associated with a diverse range of human disorders including fetal neural tube defects, age-associated dementia, infertility, and some type of cancers. Intriguingly, folate deficiency is known to cause the expression of group of rare fragile sites, all of which contain long stretch of CGG simple repeats. The most well studied locus of this kind is called FRAXA that is associated with Fragile X syndrome (FXS). Using FXS cells as a model, we demonstrated that folate deprivation triggers the extensive missegregation and aneuploidy of chromosome X [11], and MiDAS at the FRAXA locus via the break-induced DNA replication (BIR) that requires the SLX1/SLX4 endonuclease complex, the RAD51 recombinase and POLD3 [12]. Recently, based on a combination of bioinformatic and cellular biology analysis, we demonstrated that folate deficiency could cause the abnormal segregation of a region with CG-Rich trinucleotide repeats on human chromosome 2 [13]. We are currently investigating other regions that are vulnerable to folate deficiency in the human genome, and the strategies cells employ to maintain the stability of those regions (Figure 2).
References (publications from Liu group are in bold)
- Helmrich, A., M. Ballarino, and L. Tora, Collisions between Replication and Transcription Complexes Cause Common Fragile Site Instability at the Longest Human Genes. Molecular Cell, 2011. 44(6): p. 966-977.
- Letessier, A., G.A. Millot, S. Koundrioukoff, A.M. Lachages, N. Vogt, R.S. Hansen, B. Malfoy, O. Brison, and M. Debatisse, Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature, 2011. 470(7332): p. 120-3.
- Richards, R.I., Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet, 2001. 17(6): p. 339-45.
- Bjerregaard, V.A., O. Ozer, I.D. Hickson, and Y. Liu, The Detection and Analysis of Chromosome Fragile Sites. Methods Mol Biol, 2018. 1672: p. 471-482.
- Garribba, L., W. Wu, O. Ozer, R. Bhowmick, I.D. Hickson, and Y. Liu, Inducing and Detecting Mitotic DNA Synthesis at Difficult-to-Replicate Loci. Mechanisms of DNA Recombination and Genome Rearrangements: Intersection between Homologous Recombination, DNA Replication and DNA Repair, 2018. 601: p. 45-58.
- Minocherhomji, S., S. Ying, V.A. Bjerregaard, S. Bursomanno, A. Aleliunaite, W. Wu, H.W. Mankouri, H. Shen, Y. Liu, and I.D. Hickson, Replication stress activates DNA repair synthesis in mitosis. Nature, 2015. 528(7581): p. 286-90.
- Ren, L., L. Chen, W. Wu, L. Garribba, H. Tian, Z. Liu, I. Vogel, C. Li, I.D. Hickson, and Y. Liu, Potential biomarkers of DNA replication stress in cancer. Oncotarget, 2017. 8(23): p. 36996-37008.
- Wu, W., R. Bhowmick, I. Vogel, O. Ozer, F. Ghisays, R.S. Thakur, E. Sanchez de Leon, P.H. Richter, L. Ren, J.H. Petrini, I.D. Hickson, and Y. Liu, RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. Nat Struct Mol Biol, 2020. 27(5): p. 424-437.
- Wu, W., S.A. Barwacz, R. Bhowmick, K. Lundgaard, M.M. Goncalves Dinis, M. Clausen, M.T. Kanemaki, and Y. Liu, Mitotic DNA synthesis in response to replication stress requires the sequential action of DNA polymerases zeta and delta in human cells. Nat Commun, 2023. 14(1): p. 706.
- Roux, K.J., D.I. Kim, M. Raida, and B. Burke, A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. Journal of Cell Biology, 2012. 196(6): p. 801-810.
- Bjerregaard, V.A., L. Garribba, C.T. McMurray, I.D. Hickson, and Y. Liu, Folate deficiency drives mitotic missegregation of the human FRAXA locus. Proc Natl Acad Sci U S A, 2018.
- Garribba, L., V.A. Bjerregaard, M.M. Goncalves Dinis, O. Ozer, W. Wu, D. Sakellariou, J. Pena-Diaz, I.D. Hickson, and Y. Liu, Folate stress induces SLX1- and RAD51-dependent mitotic DNA synthesis at the fragile X locus in human cells. Proc Natl Acad Sci U S A, 2020. DOI: 10.1073/pnas.1921219117.
- Garribba, L., I. Vogel, M. Lerdrup, M.M. Goncalves Dinis, L. Ren, and Y. Liu, Folate Deficiency Triggers the Abnormal Segregation of a Region With Large Cluster of CG-Rich Trinucleotide Repeats on Human Chromosome 2. Front Genet, 2021. 12: p. 695124.
- Wu W, Barwacz SA, Bhowmick R, Lundgaard K, Gonçalves Dinis MM, Clausen M, Kanemaki MT, Liu, Y. Mitotic DNA synthesis in response to replication stress requires the sequential action of DNA polymerases zeta and delta in human cells. Nature Communications 2023 Feb 9;14(1):706. doi: 10.1038/s41467-023-35992-5.
- Bhowmick R, Lerdrup M, Gadi SA, Rossetti GG, Singh MI, Liu Y, Halazonetis TD, Hickson ID. RAD51 protects human cells from transcription-replication conflicts. Molecular Cell, 2022, Sep 15;82(18):3366-3381.e9. doi: 10.1016/j.molcel.2022.07.010.
- Meijering AEC, Sarlos K, Nielsen CF, Witt H, Harju J, Kerklingh E, Haasnoot GH, Bizard AH, Heller I, Broedersz CP*, Liu Y, Peterman E J G, Hickson I D*, Wuite G J L*. Nonlinear mechanics of human mitotic chromosomes. Nature 2022 605, 545-550. (*:corresponding authors)
- Schubert L, Hendriks IA, Hertz EPT, Wu W, Selles-Baiget S, Hoffmann S, Viswalingam KS, Gallina I, Pentakota S, Benedict B, Johansen J, Apelt K, Luijsterburg MS, Rasmussen S, Lisby M, Liu Y, Nielsen ML, Mailand N*, Duxin JP*. (2022). SCAI promotes error-free repair of DNA interstrand crosslinks via the Fanconi anemia pathway. EMBO Reports 23, e53639. (*:corresponding authors)
- Garribba L, Vogel I, Lerdrup M, Goncalves Dinis MM, Ren L and Liu Y. Folate Deficiency Triggers the Abnormal Segregation of a Region With Large Cluster of CG-Rich Trinucleotide Repeats on Human Chromosome 2. Frontiers in Genetics, 2021, 12:695124.
- Xing M, Zhang F, Liao H, Chen S, Che L, Wang X, Bao Z, Ji F, Chen G, Zhang H, Li W, Chen Z, Liu Y, Hickson ID*, Shen H*, and Ying S*, Replication Stress Induces ATR/CHK1-Dependent Nonrandom Segregation of Damaged Chromosomes. Molecular Cell, 2020. 78(4): p. 714-724 e5. (*:corresponding authors)
- Wu, W, Hickson ID, and Liu Y, The prevention and resolution of DNA replication–transcription conflicts in eukaryotic cells. Genome Instability & Disease, 2020. 1: p. 114–128.
- Wu W$, Bhowmick R$, Vogel I, Ozer O, Ghisays F, Thakur RS, Sanchez de Leon E, Richter PH, Ren L, Petrini JH, Hickson ID*, and Liu Y*, RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. Nature Structural & Molecular Biology 2020. 27(5): p. 424-437. ($: Joint authorship; *:corresponding authors)
- This publication was featured in SUND Insight May 2020: https://healthsciences.ku.dk/newsfaculty-news/2020/05/researchers-find-protein-that-helps-cancer-cells-to-survive/
- Garribba L, Bjerregaard VA, Goncalves Dinis MM, Ozer O, Wu W, Sakellariou D, Pena-Diaz J, Hickson ID, and Liu Y, Folate stress induces SLX1- and RAD51-dependent mitotic DNA synthesis at the fragile X locus in human cells. Proc Natl Acad Sci U S A, 2020. 117(28): p. 16527-16536.
- Pladevall-Morera D, Munk S, Ingham A, Garribba L, Albers E, Liu Y, Olsen JV, Lopez-Contreras AJ. Proteomic characterization of chromosomal common fragile site (CFS)-associated proteins uncovers ATRX as a regulator of CFS stability. Nucleic Acids Res. 2019 Sep 5;47(15):8004-8018
- Bhowmick, R, Thakur, RS, Venegas, AB, Liu, Y, Nilsson, J, Barisic, M and Hickson, ID (2019) The RIF1-PP1 axis controls abscission timing in human cells. Current Biology 2019 Apr 1;29(7):1232-1242.e5.
- Bjerregaard VA, Garribba L, McMurray CT, Hickson ID, Liu Y. Folate deficiency drives mitotic missegregation of the human FRAXA locus. Proc Natl Acad Sci USA. 2018 Dec 115 (51) 13003-13008
- Özer Ö, Bhowmick R, Liu Y, Hickson ID. Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget. 2018 Mar 9(22):15836-15846
- Garribba L*, Wu W*, Özer Ö, Bhowmick R, Hickson ID, Liu, Y. Inducing and Detecting Mitotic DNA Synthesis at Difficult-to-Replicate Loci. Methods Enzymol. 2018;601:45-58. (*: Joint authorship)
- Bjerregaard VA, Özer Ö, Hickson ID, Liu Y. The Detection and Analysis of Chromosome Fragile Sites. Methods in Molecular Biology. 2018;1672:471-482
- Ren L, Chen L, Wu W, Garribba L, Tian H, Liu Z, Vogel I, Li C, Hickson ID, Liu Y. Potential biomarkers of DNA replication stress in cancer. Oncotarget. 2017 Jun 6;8(23):36996-37008
- Kessler BM, Bursomanno S, McGouran JF, Hickson ID, and Liu Y. Biochemical and Mass Spectrometry-Based Approaches to Profile SUMOylation in Human Cells. Methods in Molecular Biology. 2017;1491:131-144
- Ghamrasni SE*, Cardoso R*, Li L, Bjerregaard VA, Liu Y, Venkatesan S, Hande MP, Henderson JT, Sanchez O, Hickson ID, Hakem A and Hakem R. Rad54 and Mus81 cooperation promotes DNA damage repair and restrains chromosome missegregation. Oncogene. 2016 Sep 15;35(37):4836-45. (*: Joint authorship)
- Nielsen CF, Huttner D, Bizard AH, Hirano S, Li TN, Palmai-Pallag T, Bjerregaard VA, Liu Y, Nigg EA, Wang LHC*, Hickson ID*. PICH promotes sister chromatid disjunction in mitosis: evidence of functional co-operation with topoisomerase II. Nature Communications, 2015 Dec. 8;6:8962. (*:corresponding authors)
- Minocherhomji S*, Ying SM*, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen HH§, Liu Y§, Hickson ID§. Replication Stress Activates DNA Repair Synthesis in Mitosis. Nature, 2015 Dec.; 528(7581):286-90. (*: joint authorship; §: corresponding authors)
- This paper was featured in ‘Research Highlight’ in Nature Review Molecular Cell Biology. 2015, doi: 10.1038/nrm.2015.24 & Nature Review Genetics. 2016, doi:10.1038/nrg.2015.35.
- This paper was recommended in F1000 Prime (http://f1000.com/prime/725980150).
- Newman JA, Savitsky P, Allerston CK, Bizard AH, Özer Ö, Sarlós K, Liu Y, Pardon E, Steyaert J, Hickson ID, Gileadi O. Crystal structure of the Bloom's syndrome helicase indicates a role for the HRDC domain in conformational changes. Nucleic Acids Research, 2015 May 26;43(10):5221-35
- Bursomanno S, McGouran JF, Kessler BM, Hickson ID, and Liu Y. Regulation of SUMO2 target proteins by the proteasome in human cells exposed to replication stress Journal of Proteome Research, 2015 Apr 3;14(4):1687-99
- Bursomanno S, Beli P, Khan AM, Minocherhomji S, Wagner SA, Bekker-Jensen S, Mailand N, Choudhary C, Hickson ID, Liu Y. Proteome-wide analysis of SUMO2 targets in response to pathological DNA replication stress in human cells. DNA Repair, 2015 (25): 84-96
- Liu Y, Nielsen CF, Yao Q, Hickson ID. The origins and processing of ultra fine anaphase DNA bridges. Current Opinion in Genetics & Development 2014 Apr 30;26:1-5
- Simandlova J, Zagelbaum J, Payne MJ, Chu WK, Shevelev I, Hanada K, Chatterjee S, Reid DA, Liu Y, Janscak P, Rothenberg E, Hickson ID. FBH1 disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. Journal of Biological Chemistry, 2013 Nov 22;288(47):34168-80
- Biebricher A, Hirano S, Enzlin JH, Wiechens N, Streicher WW, Huttner D, Wang LH, Nigg EA, Owen-Hughes T, Liu Y, Peterman E, Wuite GJ, Hickson ID., PICH: A DNA Translocase Specially Adapted for Processing Anaphase Bridge DNA. Molecular Cell, 2013 Sep 12;51(5):691-701
- Ying S*, Minocherhomji S*, Chan KL, Palmai-Pallag T, Chu WK, Wass T, Mankouri HW, Liu Y, Hickson ID. MUS81 promotes common fragile site expression. Nature Cell Biology, 2013 Aug;15(8):1001-7. doi: 10.1038/ncb2773 (*: joint authorship)
- This paper was recommended in F1000 Prime (http://f1000.com/prime/718057435).
- Wilding JL, McGowan S, Liu Y, Bodmer WF, Replication error deficient and proficient colorectal cancer gene expression differences caused by 3'UTR polyT sequence deletions. Proc Natl Acad Sci U S A. 2010 Dec 7;107(49):21058-63.
- Bracht K, Nicholls AM, Liu Y, Bodmer, WF, 5-Fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency, British Journal of Cancer, 2010, 103:340-346
- Chan CW, Wong NA, Liu Y, Bicknell D, Turley H, Hollins L, Miller CJ, Wilding JL and Bodmer WF, Gastrointestinal differentiation marker Cytokeratin 20 is regulated by homeobox gene CDX1, Proc Natl Acad Sci USA, 2009, 106(6):1936-41
- Yardy GW, Bicknell DC, Wilding JL, Bartlett S, Liu Y, Winney B, Turner GDH, Brewster SF, Bodmer WF, Mutations in the AXIN1 Gene in Advanced Prostate Cancer, European Urology, 2009, 56:486–494
- Bignone PA*, Lee KW*, Liu Y, Emilion G, Finch J, Soosay AER, Charnock FML, Beck S, Dunham I, Mungall AJ, and Ganesan TS. RPS6KA2, a putative tumour suppressor gene at 6q27 in sporadic epithelial ovarian cancer, Oncogene, 2007, 26:683–700 (*: joint authorship)
- Gaasenbeek M, Howarth K, Rowan AJ, Gorman P, Jones AA, Chaplin T, Liu Y, Bicknell D, Davison EJ, Fiegler H, Carter NP, Roylance RR, and Tomlinson IPM. Combined Array-Comparative Genomic Hybridization and Single-Nucleotide Polymorphism-Loss of Heterozygosity Analysis Reveals Complex Changes and Multiple Forms of Chromosomal Instability in Colorectal Cancers, Cancer Research, 2006, 66 (7):3471-3479
- Liu Y, and Bodmer WF. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc Natl Acad Sci USA, 2006, 103(4):976-81
- Wong NACS, Wilding J, Bartlett S, Liu Y, Warren BF, Piris J, Maynard N, Marshall R, Bodmer WF. CDX1 is an important molecular mediator of Barrett’s metaplasia, Proc Natl Acad Sci USA, 2005, 102(21):7565-7570
- Liu, Y, Bodmer, WF. Carcinogenesis of Colorectal Cancer. In Cancer De Colon, Reto E Anus, 2004, p43-54, Eds Rossi B. M. et al. Lemar Sao Paulo, Brazil
- Liu Y, Dodds P, Emilion G, Mungall AJ, Dunham I, Beck S, Wells RS, Charnock FM, Ganesan TS. The human homologue of unc-93 maps to chromosome 6q27 - characterisation and analysis in sporadic epithelial ovarian cancer. BMC Genetics, 2002, 15:3-20
- Liu Y and Ganesan TS, Tumour suppressor genes in sporadic epithelial ovarian cancer. Reproduction. 2002, 123 (3):341-53
- Liu Y, Emilion G, Mungall AJ, Dunham I, Beck S, Le Meuth-Metzinger VG, Shelling AN, Charnock FM, Ganesan TS. Physical and transcript map of the region between D6S264 and D6S149 on chromosome 6q27, the minimal region of allele loss in sporadic epithelial ovarian cancer. Oncogene. 2002, 21(3):387-99
- Yu ZY, Qi XM, Liu Y, Lu YP Yang ML. Studies of oncogenes in pregnancy gestational trophoblastic disease. Chinese Journal of Pathophysiology. 2002, 18(5):566-567 (In Chinese) 余志英 漆秀梅 刘英 卢运萍. 杨木兰, 早孕绒毛及妊娠滋养细胞疾病中癌基因表达的研究, 中国病理生理杂志 2002,18(5): 566-567
- Yu ZY, Qi XM, Liu Y, Yang ML Lu YP. A study on the expression of C-myc and N-ras in gestational trophoblastic disease. Acta Univ Med Tongji; 2001, 30 (4): 363-365, 374 (In Chinese) 余志英 漆秀梅 刘英 杨木兰. 卢运萍, 癌基因c-myc和n-ras 在妊娠滋养细胞疾病中表达的研究, 同济医科大学学报2001, 30 ( 4): 363-365, 374
- Qi XM, Liu Y. Expression of C-myc in endometrial carcinoma. Chinese Journal of Clinical Oncology, 1997, 24(8):585-589 (In Chinese) 漆秀梅 刘英 c-myc基因在子宫内膜癌及癌旁组织中表达的研究, 中国肿瘤临床1997,24(8):585-589
- Tang YP, Liu Y. Sensitivity of Biotin-labelled C—myc in in situ hybridisation. Acta Univ Med Tongji, 1996, 25 (3): 247 (In Chinese) 唐艳平 刘英 C-myc的生物素标记及灵敏度检测 华中科技大学学报(医学版) 1996, 25 (3): 247
• Dr. Cynthia T. McMurray, Berkeley National Laboratory, University of California, USA
• Prof. Chunaram Choudhary, NNF Center for Protein Research, University of Copenhagen, Denmark
• Prof. Liqun Ren, Chengde Medical University, China
• Prof. Thanos Halazonetis, University of Geneva, Switzerland
• Dr. John H. J. Petrini, Memorial Sloan Kettering Cancer Center, New York, USA
• Prof. Masato Kanemaki, Molecular Cell Engineering Laboratory, National Institute of Genetics, Yata 1111, Mishima, Shizuoka 411-8540, Japan
• Prof. Dr. Utz Fischer, University of Wuerzburg, Wuerzburg, Germany
2010 - present, teach Molecular Biology and Genetics in the first year of the Human Biology Master program, a Copenhagen Master of Excellence program (one of the 12 elite Master programs sponsored by the Danish Ministry of Science, Technology and Innovation).
Give lectures and supervise in lab and bioinformatic practical courses. Lectures include ‘Introduction to Genetics’, ‘Cancer Genetics’, ‘Introduction to Bioinformatics, and ‘Cell Cycle’. The lab course is focused on cloning and protein expression. The bioinformatic course is focused on database search.
2011 – 2015: the organiser of Health Aging IARU summer school
2015 – present: teach ‘DNA Replication, Repair and Gene Regulation’ in the Medical Cell and Tissue Biology course for first year Medicine / Odontology students.
 Group Leader
Group Leader
Ying Liu
Associate Professor
ying@sund.ku.dk
(+45) 35 32 77 61
CV, publication, etc
Molecular Aging Program
Center for Chromosome Stability
Center for Healthy Aging
