Miller R. Group
Glycosaminoglycans are clinically used heterogeneous polysaccharides drugs derived from animal sources. My lab works on applying genetic engineering of the GAG biosynthetic pathway in cells to generate materials that can be used to systematically develop instrumentation, such as mass spectrometry and single-molecule methods. Characterisation of the glycosaminoglycan sulfation code with its function paves the way for the development of sustainable lab-derived homogenous glycosaminoglycan drugs.
Glycosylation is one of the most common post-translational modifications (PTMs) of proteins essential to all eukaryotic systems, where glycans (ie. carbohydrates) are covalently attached to the protein (Fig. 1). The biological role of glycosylation in the extracellular matrix is vast, including; trafficking cells in the immune system, cell adhesion, recognizing foreign materials, controlling cell metabolism, to providing cartilage and tendon flexibility. Research and development to harness these extracellular glycomes, even when limited to the simplest forms of glycosylation, have led to the approvals of over 200 biologicals to treat diverse diseases with a global market value of €250 billion and a 10% annual growth. Glycosaminoglycan (GAG) polysaccharides is one example of these carbohydrates, they have relatively simple repeating disaccharide backbones, however, their complexity and structural diversity arises through extensive sulfation of these disaccharides (Fig. 1) and my group focus predominately on this complex molecule.
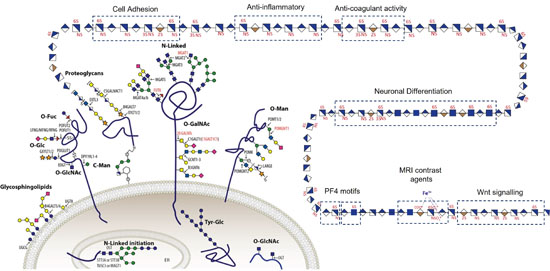
Figure 1. All cells are covered in a husk of carbohydrates including glycolipids, O- and N-linked glycans and glycosaminoglycans which is essential for all life.
Mammalian glycosaminoglycans (GAGs) are sulfated polysaccharides commonly exemplified by their most abundant forms: heparin, heparan sulfate (HS) and chondroitin/dermatan sulfate (CS/DS). GAGs are major components of the extracellular matrix that regulate many diverse functions, such as cell migration, injury, anticoagulation, angiogenesis, inflammation, as well as normal growth and development. GAGs are primarily defined by their disaccharide repeats that are extensively sulfated in distinct patterns along the chain. These sulfation codes are the source of the structural complexity and heterogeneity of GAGs, which dictate the functional properties. Despite the biological importance of GAGs, analysis continues to be challenging. Illustrating this conundrum is the widely used anti-coagulant drug heparin, a highly sulfated heparan sulfate (HS) isolated from animals, which is a heterogenous mixture lacking complete structural characterization. Whilst heparin is successful in preventing and treating thromboembolic events, a great variety of other biological effects are attributed to the heterogeneous heparin, including anti-inflammatory, anti-metastatic, and anti-viral properties. Given that the effects are expected to arise from distinct sulfation codes in GAGs, our knowledge of which codes lead to therapeutic effects is very limited due to the lack of tools to read the sulfation codes in GAGs, where my group works on the development of tools and technologies to define these sulfation code to develop homogenous GAG chains as therapeutics.
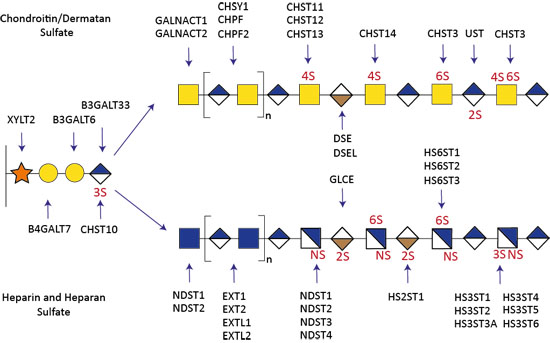
Figure 2. Genetic dissection of the GAG biosynthetic pathway.
The biosynthesis of GAGs involves a large number of glycosyltransferases and over 40 sulfotransferases. Using the KO/KI of genes orchestrating the biosynthesis of GAGs in CHO cells, we have been able to produce cell-derived libraries that display distinct heparin, HS, CS, and DS on the cell surface (Fig 2). Therefore, my groups aims to investigate how the sulfation codes lead to specific biochemical properties. The availability of this GAG library (i) advances the dissection of these sulfation codes into structure-function relationships, (ii) enables the ability to tailor the design of homogenous GAG molecules with the desired biological effects, such as anti-coagulant, anti-inflammatory or anti-viral properties, and (iii) enables the development of analytical tools and technology development, including a variety of mass spectrometry instruments and single molecule imaging modalities of GAG:protein complexes (mass photometry), and (iv) sparks new strategies for molecular dissection of GAG specific roles such as proliferation, adhesion, angiogenesis, and protein gradients (such as TGF-beta, Wnt) involved in growth and development.
-
Anggara K, Sršan L, Jaroentomeechai T, Wu X, Rauschenbach S, Narimatsu Y, Clausen H, Ziegler T, Miller RL, Kern K. Direct observation of glycans bonded to proteins and lipids at the single-molecule level. Science. 2023 Oct 13;382(6667):219-223.
- Karlsson R, Chopra P, Joshi A, Yang Z Vakhrushev SY, Clausen TM, Painter CD, Szekeres G, Chen YH, Sandoval DR, Hansen L, Esko JD, Pagel K, Dyer DP, Turnbull JE, Clausen H, Boons GJ, Miller RL. Dissecting structure-function of 3-O-sulfated heparin and engineered heparan sulfates. Sci Adv (2021), 7; 52, eabl6026.
- Szekeres GP, Krekic S, Miller RL, Mero M, Pagel K, Heiner Z. The interaction of chondroitin sulfate with a lipid monolayer observed by using nonlinear vibrational spectroscopy. Phys Chem Chem Phys (2021), 23 (23), 13389-13395.
- Zappe A, Miller RL, Struwe WB, Pagel K. State-of-the-art glycosaminoglycan characterization. Mass Spectrom Rev (2021).
- Lettow M, Greis K, Grabarics M, Horlebein J, Miller RL, Meijer G, Heldon GV, Pagel K. Chondroitin sulfate disaccharides in the gas phase: differentiation and conformational constraints. J Phys Chem Lett (2021). 125;20: 4373-4379.
- Lettow M, Grabarics M, Greis K, Mucha E, Thomas DA, Chopra P, Boons GJ, Karlsson R, Turnbull JE, Meijer G, Miller RL, Helden GV, Pagel K (2020) Cryogenic Infrared Spectroscopy Reveals Structural Modularity in the Vibrational Fingerprints of Heparan Sulfate Diastereomers. Anal Chem 92:10228-10232.
- Miller RL, Guimond SE, Schwörer R, Zubkova OV, Tyler PC, Xu Y, Liu J, Boons GJ, Grabarics M, Manz C, Hofmann J, Karlsson N, Turnbull JE, Struwe WB, Pagel K (2020) SIMMS2: Shotgun Ion Mobility Mass Spectrometry of Heparan Sulfate Saccharides. Nat Commun 11:1481-1493.
- Owen J, Thomas E, Menon J, Gray M, Gill MR, Wallington S, Skaripa-Koukelli I, Miller RL, Vallis K, Carlisle R (2019) Indium-111 after-loading of preformed HEGF-containing liposomes for molecularly targeted radionuclide delivery via ultrasound-induced cavitation. J Control Release 10:222-233.
- Rojas-Macias MA, Mariethoz J, Andersson P, Jin C, Venkatakrishnan V, Aoki NP, Shinmachi D, Ashwood C, Madunic K, Zhang T, Miller RL, Horlacher O, Struwe WB, Watanabe Y, Okuda S, Levander F, Kolarich D, Rudd PM, Wuhrer M, Kettner C, Packer NH, Aoki-Kinoshita KF, Lisacek F, Karlsson NG (2019) Towards a standardized bioinformatics infrastructure for N- and O-glycomics. Nat Commun 22:3275-3285
- Roworth AP, Carr SM, Liu G, Barczak W, Miller RL, Munro S, Kanapin A, Samsonova A, La Thangue NB (2019) Arginine methylation expands the regulatory mechanisms and extends the genomic landscape under E2F control. Sci Adv 26:4640-4653.
- Miller RL, Guimond SE, Prescott M, Karlsson N, Turnbull JE (2017). Versatile Separation and Analysis of Heparan Sulfate Oligosaccharides using Graphitised Carbon Liquid Chromatography and Electrospray Mass Spectrometry. Anal Chem 5:8942-8950.
- Gray CJ, Sánchez-Ruíz A, Šardzíková I, Ahmed YA, Miller RL, Reyes Martinez JE, Pallister E, Huang K, Both P, Hartmann M, Roberts HN, Šardzík R, Mandal S, Turnbull JE, Eyers CE, Flitsch SL (2017). Label-Free Discovery Array Platform for the Characterisation of Glycan Binding Proteins and Glycoproteins. Anal Chem 18:4444-4451
- Miller RL, Guimond SE, Blocksidge J, Shivkumar M, Austin J, Leary JA, Turnbull JE (2016). Heparin isomeric oligosaccharide separation using volatile salt strong anion exchange chromatography. Anal Chem 6:11542-11550.
- Miller RL, Dykstra A, Wei W, Hosclaw C, Turnbull JE, Leary JA (2016). Enrichment of two isomeric heparin oligosaccharides with different Kd affinities towards MCP-1. Anal Chem 6:11551-11558.
- Lawless C, Holman SW, Brownridge P, Lanthaler K, Harman VM, Watkins R, Sims PFG, Miller RL, Grant CM, Eyers CE, Beynon RJ, Hubbard SJ. (2016). Absolute quantification of 1000 yeast proteins provide insight into translational control. Mol Cell Proteomics 15:1309-1322.
- Miller RL, Wei W, Turnbull JE, Leary JA (2015). Composition, sequencing and ion mobility mass spectrometry of heparan sulfate-like octasaccharide isomers differing in glucuronic and iduronic acid content. Eur J Mass Spectrom. 21:245-54.
- Wei W, Miller RL, Ninonuevo M, Leary JA (2013). Compositional analysis of heparan sulfate from in pre-menopausal and post-menopausal women indicate biosynthetic pathway changes. Anal Chem 85:5917–5923.
- Turnbull JE, Miller RL, Ahmed Y, Puvirajesinghe T, and Guimond SE (2010). Glycomic profiling of heparan sulfate structure and activity. Meth Enzymol 480:65-85
- Wagstaff SC, Favreau P, Cheneval O, Laing GD, Wilkinson MC, Miller RL, Stocklin R, Harrison RA (2008). Molecular characterisation of endogenous snake venom metalloproteinase inhibitors. Biochem Biochem Biophys Res Commun 365:650-6.
We are always looking for motivated scientist to conduct PhD or postdoc research. Please send a CV and cover letter to rmiller@sund.ku.dk
We also host BSc, MSc and ERASMUS students for research projects
- Michael Davies (Copenhagen)
- Christine Chuang (Copenhagen)
- Kevin Pagel (Germany)
- Weston Struwe (Oxford)
Novo Nordisk Foundation Hallas Moller
Carlsberg Foundation
EU H2020
Group Leader
 Rebecca Miller
Rebecca Miller
associate professor
rmiller@sund.ku.dk
(+45) 35 33 18 41
CV, publications, etc
