Miller T. Group
Our lab aims to understand the molecular mechanisms that maintain eukaryotic genome stability during DNA replication using a combination of genetic, biochemical, cellular, and structural techniques, including cryo-EM
The molecular mechanisms of faithful genome replication
Accurate chromosome replication is essential for controlled cell proliferation and the faithful transfer of genetic information from one generation to the next. A failure to maintain genome stability can cause diverse developmental disorders and age-related diseases, including accelerated neurodegeneration and cancer. Eukaryotic genomes are duplicated by large, multi-component protein machines called replisomes. At their core, replisomes consist of a helicase that unwinds duplex DNA and polymerases that catalyse the synthesis of new DNA molecules according to the parental template. A diverse range of accessory factors are also required facilitate the faithful replication of eukaryotic chromosomes, which contain an array of ‘obstacles’ that could otherwise stall or inhibit the replication machinery. Together, the replisome and accessory factors regulate the timing, speed and accuracy of replication, minimising replication errors and preventing genome instability. Our group aims to understand the molecular mechanisms by which eukaryotic replisomes and accessory factors preserve genome stability during DNA replication and how failures in these mechanisms cause disease. Ultimately, we plan to use the knowledge we gain to develop novel therapeutic approaches to prevent or treat human disorders linked to genome instability.
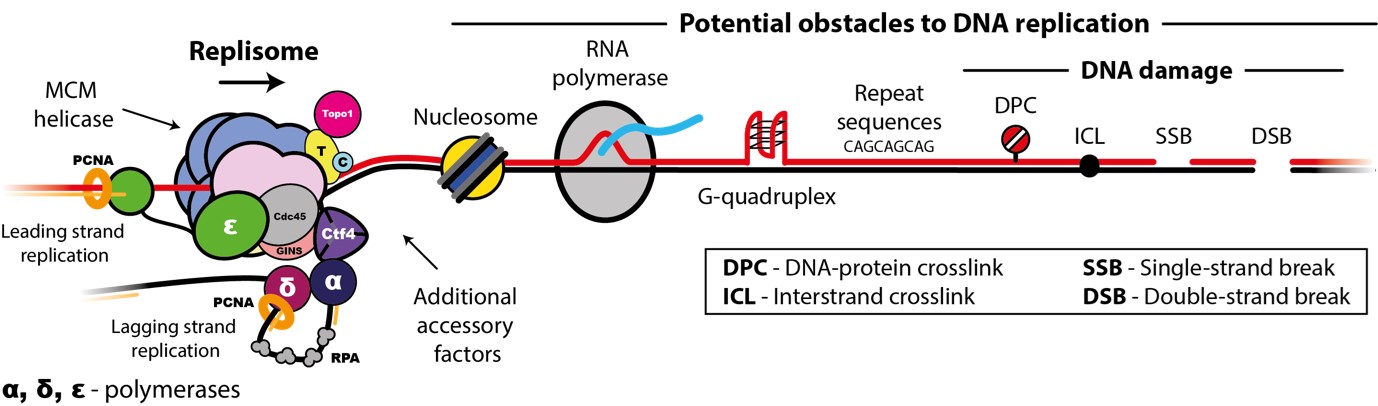
- The mechanisms and regulation of MCM helicase loading
- Replication-coupled DNA-protein crosslink (DPC) repair
- Accessory helicases in DNA replication
- Electron microscopy methods development – ‘Reconstitution in silico’ (RECONSIL)
We welcome applications from motivated postdoctoral and predoctoral researchers as well as master students. Please contact us at tmiller@sund.ku.dk
- Lim CT, Miller TCR, Tan KW, Talele S, Early A, East P, Sánchez H, Dekker NH, Costa A, Diffley JFX, Cell cycle regulation has shaped replication origins in budding yeast Nat Struct Mol Biol. 2025 Sep;32(9):1697-1707
- Sellés-Baiget S, Ambjørn SM, Carli A, Hendriks IA, Gallina I, Davey NE, Benedict B, Zarantonello A, Gadi SA, Meeusen B, Hertz EPT, Slappendel L, Semlow D, Sturla S, Nielsen ML, Nilsson J, Miller TCR, Duxin JP, Catalytic and noncatalytic functions of DNA polymerase kappa in translesion DNA synthesis Nat Struct Mol Biol. 2025 Feb;32(2):300-314
- Polasek-Sedlackova H, Miller TCR, Krejci J, Rask MB, Lukas J Solving the MCM paradox by visualizing the scaffold of CMG helicase at active replisomes. Nature Communications. 2022 Oct 14;13(1):6090.
-
Pühringer T, Greiwe JF, Miller TCR*, Costa A*. ReconSil: An electron microscopy toolbox to study helicase function at an origin of replication. Methods in Enzymology, Academic Press, 2022, ISSN 0076-6879, *Corresponding authors
-
Greiwe JF, Zanetti, G, Miller TCR*, Costa A*.“In silico reconstitution of DNA replication. Lessons from single-molecule imaging and cryo-tomography applied to single-particle cryo-EM. Current Opinion in Structural Biology, 2022, Jan 10; 72, 279-286. *Corresponding authors.
-
Greiwe JF, Miller TCR, Locke J, Martino F, Howell S, Schreiber A, Nans A, Diffley JFX, Costa A. Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase. Nature Structural and Molecular Biology, 2021 Dec 28.
- Miller TCR, Locke J, Greiwe JF, Diffley JFX, Costa A. Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature, 2019 Nov 20; 575, 704-710.
- Miller TCR, Costa A. The architecture and function of the chromatin replication machinery. Current Opinion in Structural Biology, 2017 Dec 1; 47, 9-16.
Group Leader
 Tom Miller
Tom Miller
Associate Professor of Genome Stability
tmiller@sund.ku.dk
(+45) 35 32 70 74
