Ochs Group
Our laboratory investigates the molecular mechanisms of genome stability maintenance in the 3D chromatin space. We aim to unravel how gene regulation, DNA repair, and cell division are coordinated in space and time, and how disruptions in these processes contribute to pathological conditions such as infertility, neurodevelopmental disorders, and cancer.
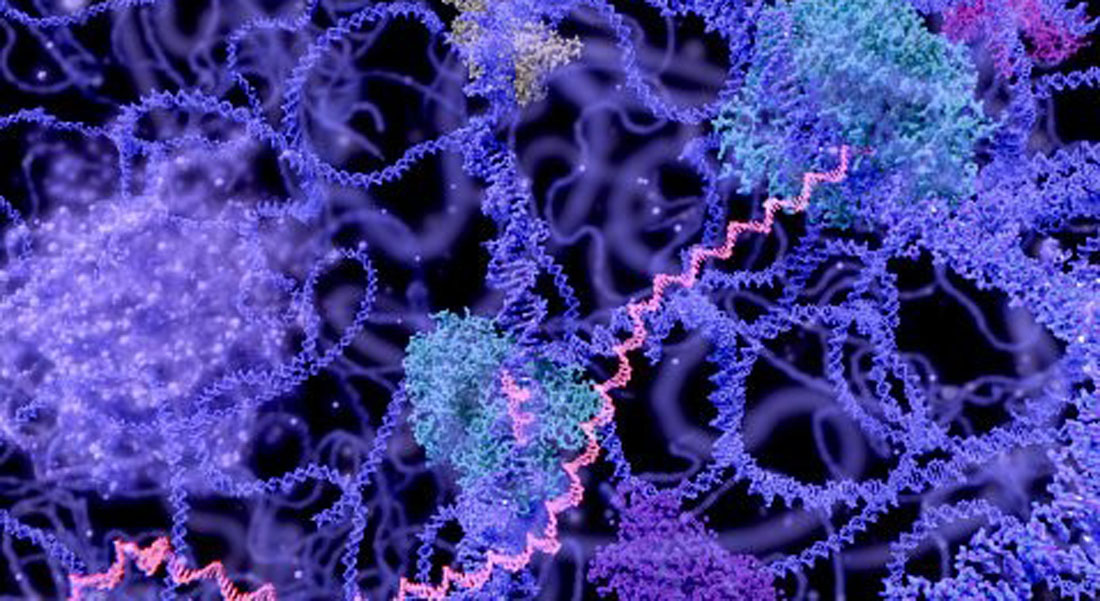
We are fascinated by the intricate three-dimensional organization of the genome within the cell nucleus and investigate how essential biological processes are orchestrated within this spatial framework. Our research focuses on the organisation of 3D chromatin, with particular emphasis on the cohesin complex. We are especially interested in cohesin’s dual roles: organizing chromatin through loop extrusion to regulate gene expression, and mediating sister chromatid cohesion to ensure accurate genome inheritance.
In parallel, we explore how 3D chromatin organization contributes to the fidelity of DNA repair. We are particularly focused on the molecular mechanisms of DNA repair and its dynamic interplay with chromatin architecture. Our previous work revealed that the 3D chromatin environment surrounding a DNA lesion plays a critical role in ensuring accurate repair. Emerging evidence suggests that DNA damage not only affects the local DNA sequence but also perturbs the broader chromatin landscape, with downstream consequences for gene expression at the affected locus. Understanding how DNA repair is coordinated with other chromatin-associated processes remains a central focus of our research.
To dissect these complex mechanisms with unprecedented spatiotemporal resolution, we develop and apply cutting-edge super-resolution imaging technologies, including the first super-resolution microscopy approach for single-molecule imaging in intact cells and on 3D chromatin.
Our research is highly interdisciplinary and collaborative, operating at the interface of molecular biology, physics, and medical sciences. We are committed to advancing fundamental understanding in chromosome biology and uncovering how perturbations in 3D chromatin-associated processes contribute to pathological conditions such as infertility, cancer, and neurodevelopmental syndromes.
Molecular mechanism of chromatin organisation by cohesin loop extrusion
Our genetic blueprint, the DNA, is intricately organized into three-dimensional chromatin structures within the cell nucleus, a spatial configuration essential for regulating gene expression, DNA repair, and replication. A key architect of this 3D chromatin landscape is the cohesin complex, a multiprotein assembly within the Structural Maintenance of Chromosomes (SMC) family. In vitro studies have suggested that cohesin functions as a molecular motor, extruding chromatin loops to shape genome architecture.
Our research aims to visualize and dissect cohesin-mediated loop extrusion within intact cells and native chromatin environments. We have recently identified two distinct cohesin populations in human cells, each with unique stoichiometries and functions (Ochs et al., Science, 2024). While cohesin involved in sister chromatid cohesion exists as a monomer, all other cohesin complexes appear multimeric.
To investigate these mechanisms, we integrate advanced gene editing with a suite of cutting-edge super-resolution microscopy techniques, including 3D Structured Illumination Microscopy (3D-SIM), DNA-PAINT, and chromatin expansion microscopy. Through these approaches, we aim to define the stoichiometry of loop extruding cohesin, uncover its regulatory determinants, and elucidate its impact on gene expression during development and cellular homeostasis.
Spatiotemporal coordination of sister chromatid cohesion
Cell division is the foundation of life, responsible for constructing our tissues, organs, and entire bodies. Each cell division is a delicate process, ensuring the accurate distribution of our genetic code from one mother cell to two daughter cells. This critical task is orchestrated by a group of proteins known as the cohesin complex.
Cohesin holds together newly duplicated sister chromatids from their genesis during DNA replication until their segregation during mitosis, in a process called sister chromatid cohesion. Despite its discovery nearly three decades ago, the molecular mechanism underlying sister chromatid cohesion remains incompletely understood. Utilizing single molecule super-resolution imaging in human cells, we have recently discovered the nature of cohesin mediating sister chromatid cohesion. Monomers of cohesin, in conjunction with an accessory factor called Sororin, entrap identical chromosomes and facilitate their symmetrical distribution to daughter cells (Ochs et al., Science, 2024). Building on this work, we are now investigating the molecular mechanism of cohesion establishment during DNA replication as well as venturing into the most relevant model system to understand the mechanism of sister chromatid cohesion – meiosis. We are specifically interested in the intersection of sister chromatid cohesion and fertility, as well as in the coordination of sister chromatid cohesion and loop extrusion to organise chromosomes in 3D.
Coupling of 3D chromatin organisation and DNA repair
We have recently discovered that there is a two-way interconnection between the processes of DNA repair and 3D chromatin organisation in eukaryotic cells (Ochs et al., Nature, 2019), and mutations in genes essential for either of these processes predispose patients to DNA damage disorders. It is so far not understood how DNA repair and 3D chromatin organisation are co-regulated and how genomic and epigenomic stability are maintained in the 3D chromatin space. With our recent development of the first super-resolution microscopy technique to study cellular processes at single molecule resolution on 3D chromatin and in intact cells (Ochs et al., Science, 2024), we can now resolve this long-standing question. By combination of multiplexed gene editing, chromatin topology analysis, and artificial intelligence-driven biomedical image analysis, we investigate how 3D chromatin contributes to faithful DNA repair, what impact DNA damage has on related processes such as gene expression, and why dysregulation of these processes drives disease. To this end, we are collaborating with Rigshospitalet, the Danish national hospital, to study the aetiology of Cornelia-de-Lange Syndrome, a so far untreatable neurodevelopmental disorder.
- Ochs, F., Green, C., Szczurek, A.T., Pytowski, L., Kolesnikova, S., Brown, J., Gerlich, D.W., Buckle, V., Schermelleh, L., Nasmyth, K.A. Sister chromatid cohesion is mediated by individual cohesin complexes. Science 383 (6687), 1122-1130 (2024). PMID: 38452070. Open access: https://www.science.org/stoken/author-tokens/ST-1749/full
- Groelly, F. J., Dagg, R. A., Petropoulos, M., Rossetti, G. G., Prasad, B., Panagopoulos, A., Paulsen, T., Karamichali, A., Jones, S. E., Ochs, F., Dionellis, V. S., Puig Lombardi, E., Miossec, M. J., Lockstone, H., Legube, G., Blackford, A. N., Altmeyer, M., Halazonetis, T. D. & Tarsounas, M. Mitotic DNA synthesis is caused by transcription-replication conflicts in BRCA2-deficient cells. Molecular Cell 82, 1-16 (2022). PMID: 36002001
- Ercilla, A., Benada, J., Amitash, S., Zonderland, G., Baldi, G., Somyajit, K., Ochs, F., Constanzo, V., Lukas, J. & Toledo, L. Physiological tolerance to ssDNA enables strand uncoupling during DNA replication. Cell Reports 30 (7), 2416-2429 (2020). PMID: 32075739
- Ochs, F., Karemore, G., Miron, E., Brown, J., Sedlackova, H., Rask, M.-B., Lampe, M., Buckle, V., Schermelleh, L., Lukas, J. & Lukas, C. Stabilization of chromatin topology safeguards genome integrity. Nature 574, 571-574 (2019). PMID: 31645724
- Somyajit, K., Gupta, R., Sedlackova, H., Neelsen, K.J., Ochs, F., Rask, M.-B., Choudhary, C. & Lukas, J. Redox-sensitive alteration of replisome architecture safeguards genome integrity. Science 358 (6364), 797-802 (2017). PMID: 29123070
- Sotiriou, S. K., Kamileri, I., Lugli, N., Evangelou, K., Da-Ré, C., Huber, F., Padayachy, L., Tardy, S., Nicati, N. L., Barriot, S., Ochs, F., Lukas, C., Lukas, J., Gorgoulis, V. G., Scapozza, L. & Halazonetis, T. D. Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Molecular Cell 46 (6), 1127-1134 (2016). PMID: PMC5179496
- Ochs, F., Somyajit, K., Altmeyer, M., Rask, M.-B., Lukas, J. & Lukas, C. 53BP1 fosters fidelity of homology-directed DNA repair. Nature Structural and Molecular Biology 23 (8), 714-721 (2016). PMID: 27348077
Group Leader
 Fena Ochs
Fena Ochs
Associate Professor
fenaochs@sund.ku.dk
(+45) 35 33 86 22
CV, publication, etc
Molecular Aging Program
The Fena Ochs Laboratory external website

The laboratory has been awarded the LEAF - Laboratory Efficiency Assessment Framework Gold certificate.





